Exploding Architectures: Dizzying Array of New Chip Designs May Spell the End of Exascale
FPGA Designs Enable 100x Speedups for Key Applications

Oak Ridge National Laboratory (ORNL) is the new home of the world’s fastest computer, Summit – arguably the first “almost exascale” computer clocking in at over 200 petaflops of calculation horsepower (able to perform 200 quadrillion floating point operations per second). But ORNL’s head of Advanced Computing, Jeff Nichols, said over the summer that “in a few years we won’t be talking about exascale any more.” With governments around the world just now pouring billions into exascale computing programs, what could Nichols be talking about?
One explanation comes from looking at how specialized hardware architectures, like GPUs, FPGAs and ASICs, are achieving amazing performance improvements for specific types of calculation, such as Fast Fourier Transforms, a mathematical operation on time series data that is fundamental to many analysis and simulation techniques. Massive speedup of FFT and similar operations could enable game-changing performance improvements for key analysis and modelling applications – shorten times to solution for large categories of DI users, and reduce the demand for general purpose exascale systems like Summit.
This is not science fiction – it’s been happening for years. Here are some examples:

Edico Genome has created an FPGA-based system called DRAGEN that speeds up genomic processing by up to 100x – providing results in 30 minutes instead of 30 hours. Since genomics is one of the fastest growing drivers of demand for digital infrastructure, systems like this can make life easier for any DI provider, and change the competitive landscape for more general purpose HPC solutions. The San Diego Supercomputer Center (SDSC) installed a DRAGEN system as part of its “long tail” supercomputer, COMET.
Edico was purchased earlier in 2018 by Illumina , the dominant global vendor of high-throughput DNA sequencing systems. Since sequencing systems all require purchasers to establish significant digital infrastructure to support timely storage and processing of their output data, Illumina’s purchase of Edico might limit researcher flexibilty in analysis approaches at some point in the future. Luckily several DI vendors are also building FPGA-based solutions in this area, so we should expect more “appliance” type offerings for genomic analysis.
Like Edico, Ryft (now part of BlackLynx) combines FPGAs and optimized software drivers to accelerate data analytics performance by up to 100x, particularly when searching large, unindexed datasets such as security logs, textual data, and XML files, as well as streaming datasets from security and financial monitoring systems – or scientific data sets.
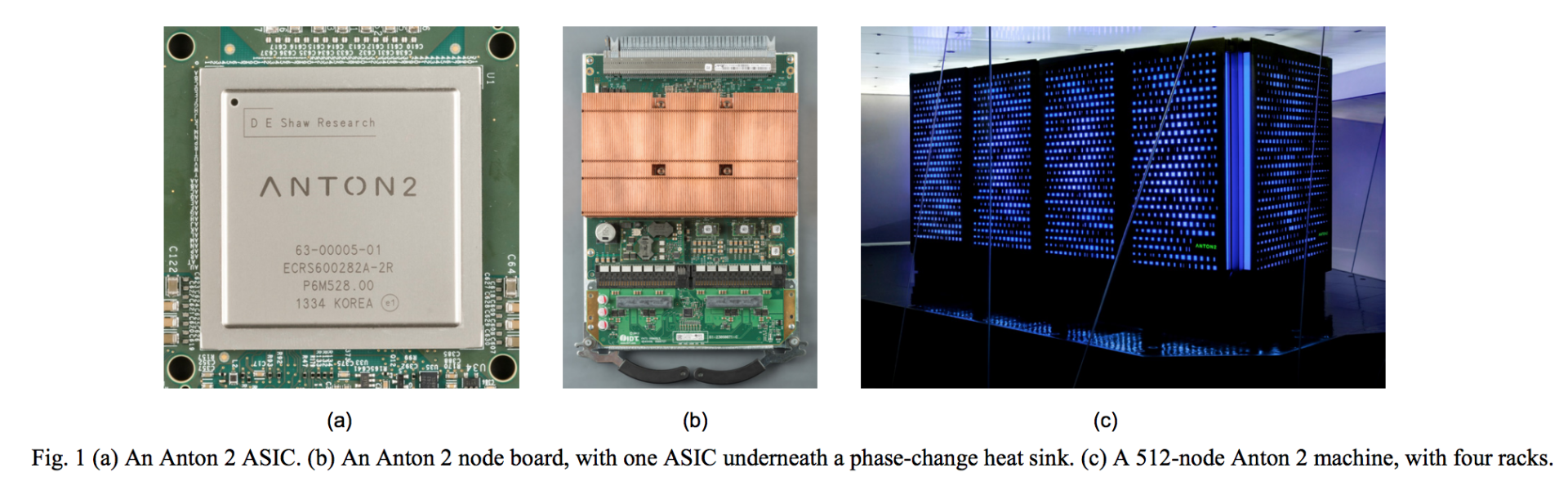
Back in 2008, DE Shaw Research created special purpose systems for molecular dynamics modelling called Anton , that provided 100x faster performance than comparably-sized HPC systems at the time. Similar to Edico and Ryft, Anton accelerated performance by using ASICs (application specific integrated circuits) that had been custom fabricated to execute the most demanding mathematical operations – using hardware instead of software -- needed for the molecular dynamics software. One Anton system was donated by DE Shaw to the Pittsburgh Supercomputer Centre (PSC). In 2016 it was replaced by a larger system, Anton 2 , that is available to academic researchers on a merit basis. Journal articles resulting from the research enabled by both Anton and Anton 2 are listed on PSC’s website .
With growing numbers of researchers using DI techniques in their work, appliance-like solutions can offer “petascale” performance to many of those researchers without the “petascale” price tag. Whether deployed in the cloud or in your data centre, this can be a game-changer for both corporate R&D and academic research and innovation.